
PhD, AFHEA
Lecturer
- About
-
- Email Address
- marius.wenzel@abdn.ac.uk
- Telephone Number
- +44 (0)1224 273206
- Office Address
- School/Department
- School of Biological Sciences
Biography
since 12/2019: Research Fellow (University of Aberdeen)
2018-2019: Bioinformatician (CGEBM, University of Aberdeen)
2015-2018: Post-doctoral research fellow (University of Aberdeen)
2011-2015: PhD Biological Science (University of Aberdeen)
2008-2011: BSc (Hons.) Biology, 1st class (University of Aberdeen)Qualifications
- AFHEA Associate Fellow of Advance HE2019 - Advance HE
- PhD Biological Science2015 - University of Aberdeen
- BSc Hons. Biology2011 - University of Aberdeen
- Research
-
Research Overview
I am an evolutionary biologist with broad interests in fundamental questions regarding the evolution of phenotypes, physiology, behaviour, ecology and biodiversity, spanning all levels of biological organisation from cells to ecosystems. The main themes of my research include phylogenomics, comparative genomics, transcriptomics, epigenomics and conservation genetics. I am particularly interested in genome evolution and the role of epigenetics in the evolution of functional phenotypic diversity.
My research emphasises the use of high-throughput sequencing to characterise genomes, transcriptomes and epigenomes in wild and laboratory systems, and involves a large amount of bioinformatics data mining. A large component of my computational interest is the utility of long-read sequencing using Oxford NanoPore and PacBio technology for characterising environmental samples, obtaining full-length transcript repertoires and assembling complex eukaryotic genomes.
Research Areas

Biological and Environmental Sciences
Research Specialisms
- Evolution
- Bioinformatics
- Genomics
- Biomolecular Science
- Transcriptomics
Our research specialisms are based on the Higher Education Classification of Subjects (HECoS) which is HESA open data, published under the Creative Commons Attribution 4.0 International licence.
Current Research
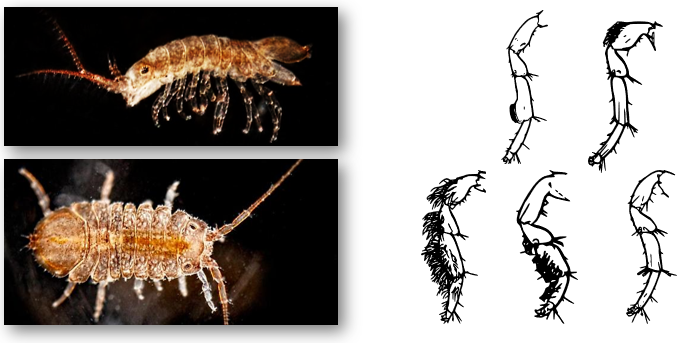
Speciation genomics in intertidal isopods (Jaera albifrons)
The Jaera albifrons species complex comprises five species that are reproductively isolated via subtle differences in male leg morphology and female preference for tactile stimulation by the male's legs during courtship.
My work aims to examine genome-wide sequence variation among the species to understand the evolutionary history and the underlying genomic architecture of reproductive isolation within the species complex. A particular exciting focus is to identify candidate genes that are functionally involved in leg development and/or courtship behaviour and manipulate these genes in vivo to generate novel species diversity.
Functional behavioural genomics in the slime mould Physarum polycephalum
The slime mould Physarum polycephalum is a single-celled eukaryotic organism that forms a large amorphous blob termed a plasmodium. The plasmodium can grow indefinitely and contains millions of nuclei that govern complex problem-solving behaviour in response to environmental conditions, famously finding the most efficient path through a maze or replicating efficient national rail or road networks.
I am investing the fundamental functional genomic mechanisms responsible for driving this cognitive behaviour. The main focus is on epigenetic patterns that may affect gene expression and confer cellular memory. These insights are achieved via experimental trials and direct manipulation of the cellular epigenetic machinery.

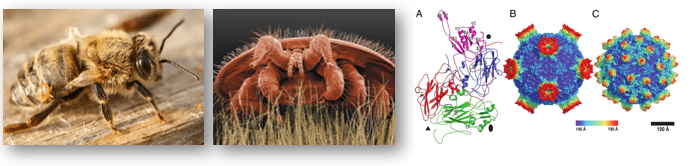
Host-parasite functional genomics in the honey-bee/Varroa/DWV symbiosis
Honey bees are experiencing a drastic decline with serious consequences for pollination rates in the wild and agricultural food security. One important cause for the decline is the parasitic mite Varroa destructor which transmits deadly viruses such as the deformed-wing virus (DWV).
My research considers the functional genomics basis of this intricate three-way symbiosis with particular focus on epigenetic factors affecting host-parasite interactions and virus transmission.
Supervision
Rosalin Simpson, QUADRAT DTP: "Harnessing Comparative and Functional Genomics to Explore Adaptive Potential in Predatory and Pest Mite Species"
Anastasia Leligdowicz, EASTBIO DTP: "Using functional genomics and epigenetics to understand the molecular underpinnings of complex behaviour in non-neural organisms"
Anna Wawer, SUPER DTP: "Immunogenomic status of salmonids in Scotland"
- Publications
-
Page 1 of 1 Results 1 to 32 of 32
Neurometabolic adaptations to intestinal inflammation in a mouse model of colitis
American journal of physiology. Gastrointestinal and liver physiologyContributions to Journals: ArticlesEphemeral signatures of phylosymbiosis in dermal microbiomes within the requiem shark family (Carcharhinidae)
Working Papers: Preprint Papers- [ONLINE] DOI: https://doi.org/10.1099/acmi.0.001140.v1
Cap-adjacent 2’-O-ribose methylation of RNA in C. elegans is required for postembryonic growth and germline development in the presence of the decapping exonuclease EOL-1
Working Papers: Preprint Papers- [ONLINE] DOI: https://doi.org/10.1101/2025.03.10.638824
A nematode-specific ribonucleoprotein complex mediates interactions between the major nematode spliced leader snRNP and its target pre-mRNAs
Nucleic Acids Research, vol. 52, no. 12, pp. 7245-7260Contributions to Journals: ArticlesMacroalgal eukaryotic microbiome composition indicates novel phylogenetic diversity and broad host spectrum of oomycete pathogens
Environmental Microbiology, vol. 26, no. 6, e16656Contributions to Journals: Articles- [ONLINE] DOI: https://doi.org/10.1111/1462-2920.16656
- [OPEN ACCESS] http://aura.abdn.ac.uk/bitstreams/5f30b0d2-4df0-461b-b992-eb43e8b2ba10/download
- [ONLINE] View publication in Scopus
Large effective population size masks population genetic structure in Hirondellea amphipods within the deepest marine ecosystem, the Mariana Trench
Molecular Ecology, vol. 32, no. 9, pp. 2206-2218Contributions to Journals: ArticlesThe transcriptomic signature of physiological trade-offs caused by larval overcrowding in Drosophila melanogaster
Insect Science, vol. 30, no. 2, pp. 539-554Contributions to Journals: Articles- [ONLINE] DOI: https://doi.org/10.1111/1744-7917.13113
- [ONLINE] View publication in Scopus
A novel, essential trans-splicing protein connects the nematode SL1 snRNP to the CBC-ARS2 complex
Nucleic Acids Research, vol. 50, no. 13, pp. 7591-7607Contributions to Journals: ArticlesMonodopsis and Vischeria genomes shed new light on the biology of eustigmatophyte algae
Genome biology and evolution, vol. 13, no. 11, evab233Contributions to Journals: ArticlesSLIDR and SLOPPR: flexible identification of spliced leader trans-splicing and prediction of eukaryotic operons from RNA-Seq data
BMC Bioinformatics, vol. 22, 140Contributions to Journals: ArticlesComparative genomics of cetartiodactyla: energy metabolism underpins the transition to an aquatic lifestyle
Conservation Physiology, vol. 9, no. 1, coaa136Contributions to Journals: ArticlesResolution of polycistronic RNA by SL2 trans-splicing is a widely-conserved nematode trait
RNA , vol. 26, no. 12, pp. 1891-1904Contributions to Journals: Articles- [ONLINE] DOI: https://doi.org/10.1261/rna.076414.120
- [ONLINE] View publication in Scopus
Deep evolutionary origin of nematode SL2 trans-splicing revealed by genome-wide analysis of the Trichinella spiralis transcriptome
bioRxivContributions to Journals: Articles- [ONLINE] DOI: https://doi.org/10.1101/642082
- [ONLINE] http://biorxiv.org/cgi/content/short/642082v1?rss=1
- [ONLINE] View publication in Mendeley
Parasite-mediated selection in red grouse--consequences for population dynamics and mate choice
Wildlife Disease Ecology: Linking Theory to Data and Application. Wilson, K., Fenton, A., Tompkins, D. (eds.), pp. 296-320Chapters in Books, Reports and Conference Proceedings: ChaptersAdaptations of energy metabolism in cetaceans have consequences for their response to foraging disruption
Working Papers: Preprint Papers- [ONLINE] DOI: https://doi.org/10.1101/709154
Microbiome composition within a sympatric species complex of intertidal isopods (Jaera albifrons)
PloS ONE, vol. 13, no. 8, e0202212Contributions to Journals: ArticlesSignatures of balancing selection in toll-like receptor (TLRs) genes – novel insights from a free-living rodent
Scientific Reports, vol. 8, 8361Contributions to Journals: Articles- [ONLINE] DOI: https://doi.org/10.1038/s41598-018-26672-2
- [OPEN ACCESS] http://aura.abdn.ac.uk/bitstreams/7940ccc8-776e-4cad-b6f8-a418909c69fc/download
- [ONLINE] View publication in Scopus
Phylogenomic signatures of speciation and phylogeography in a species complex of sympatric intertidal isopods (Jaera albifrons).
Evolution 2018: Joint congress on Evolutionary BiologyContributions to Conferences: PostersRADseq-based phylogenomics fully resolves evolutionary relationships in a species complex of sympatric intertidal isopods (Jaera albifrons).
European Society of Evolutionary Biology (ESEB) XVI CongressContributions to Conferences: PostersThe genomic landscape of pre-zygotic reproductive isolation in a sympatric species complex of intertidal isopods (Jaera albifrons)
Contributions to Conferences: Oral PresentationsThe role of parasite-driven selection in shaping landscape genomic structure in red grouse (Lagopus lagopus scotica)
Molecular Ecology, vol. 25, no. 1, pp. 324-341Contributions to Journals: Articles- [ONLINE] DOI: https://doi.org/10.1111/mec.13473
- [OPEN ACCESS] http://aura.abdn.ac.uk/bitstreams/a58c5d0b-b647-4751-b126-ff9f5b4ba908/download
Genome-wide association and genome partitioning reveal novel genomic regions underlying variation in gastrointestinal nematode burden in a wild bird
Molecular Ecology, vol. 24, no. 16, pp. 4175-4192Contributions to Journals: Articles- [ONLINE] DOI: https://doi.org/10.1111/mec.13313
- [OPEN ACCESS] http://aura.abdn.ac.uk/bitstreams/72ad4f2f-137d-453a-b929-c13fdabc7dca/download
In silico identification and characterisation of 17 polymorphic anonymous non-coding sequence markers (ANMs) for red grouse (Lagopus lagopus scotica)
Conservation Genetics Resources, vol. 7, no. 2, pp. 319-323Contributions to Journals: ArticlesDigging for gold nuggets: uncovering novel candidate genes for variation in gastrointestinal nematode burden in a wild bird species
Journal of Evolutionary Biology, vol. 28, no. 4, pp. 807-825Contributions to Journals: Articles- [ONLINE] DOI: https://doi.org/10.1111/jeb.12614
- [OPEN ACCESS] http://aura.abdn.ac.uk/bitstreams/624ecdba-7e37-41cd-bcc2-0cd6fae0bef5/download
Identification and characterisation of 17 polymorphic candidate genes for response to parasitic nematode (Trichostrongylus tenuis) infection in red grouse (Lagopus lagopus scotica)
Conservation Genetics Resources, vol. 7, no. 1, pp. 23-28Contributions to Journals: ArticlesFine-scale epigenetic structure in relation to gastrointestinal parasite load in red grouse (Lagopus lagopus scotica).
European Society for Evolutionary Biology (ESEB) XV CongressContributions to Conferences: PostersLandscape genomics patterns in red grouse (Lagopus lagopus scotica) – a role for parasites?
Contributions to Conferences: Oral PresentationsFine-scale population epigenetic structure in relation to gastrointestinal parasite load in red grouse (Lagopus lagopus scotica)
Molecular Ecology, vol. 23, no. 17, pp. 4256-4273Contributions to Journals: Articles- [ONLINE] DOI: https://doi.org/10.1111/mec.12833
- [OPEN ACCESS] http://aura.abdn.ac.uk/bitstreams/1ce6895b-8dbb-4168-bbe0-761aa6b1531d/download
A transcriptomic investigation of handicap models in sexual selection
Behavioral Ecology and Sociobiology, vol. 67, no. 2, pp. 221-234Contributions to Journals: ArticlesTools and approaches for disentangling the effects of parasite-driven processes on host neutral and adaptive genetic diversity
European Society for Evolutionary Biology (ESEB) XIV CongressContributions to Conferences: PostersPronounced genetic structure and low genetic diversity in European red-billed chough (Pyrrhocorax pyrrhocorax) populations
Conservation Genetics, vol. 13, no. 5, pp. 1213-1230Contributions to Journals: ArticlesIsolation and characterisation of 17 microsatellite loci for the red-billed chough (Pyrrhocorax pyrrhocorax)
Conservation Genetics Resources, vol. 3, no. 4, pp. 737-740Contributions to Journals: Articles
