
Senior Lecturer
- About
-
- Email Address
- a.lionikas@abdn.ac.uk
- Telephone Number
- +44 (0)1224 438025
- Office Address
School of Medicine, Medical Sciences and Nutrition
IMS Building, Room 2:21
University of Aberdeen Foresterhill Aberdeen AB25 2ZD
- School/Department
- School of Medicine, Medical Sciences and Nutrition
Biography
I am senior lecturer at the school of Medicine, Medical Sciences and Nutrition, teaching topics related to physiology, genetics, and sports science. I also conduct research aiming to understand the mechanisms underlying the morphology and function of skeletal muscle, with a particular focus on the role of genetic factors. Skeletal muscle is responsible for vital functions such as locomotion, respiration, and maintenance of glucose homeostasis and its decline due to disease or aging presents an increasing healthcare concern. As the results of the work in my lab, we know now that there are hundreds of genes affecting skeletal muscle and the work to identify their role and mechanisms is ongoing.
My career has been significantly influenced by interests in sports and exercise. I graduated from the Lithuanian Sports University as a PE teacher in 1992, received a Masters degree in 1994 and was awarded a PhD from the same institution in 1999 for the work on acute muscle adaptation to exercise. From 1999 to 2000, I was a Guest Researcher at the Department of Physiology and Pharmacology at the Karolinska Institute (advisor prof Jan Henriksson) in Sweden working on the effects of exercise on muscle signalling. From 2001 to 2003, I did postdoctoral work in muscle biology at the Pennsylvania State University (USA) and Uppsala University (Sweden; advisor prof Lars Larsson). After my second postdoctoral position in quantitative genetics at Pennsylvania State University (advisors Dr David A. Blizard and Dr George P. Vogler; from 2003 to 2007) I continued to work there as a Research Associate until 2009. In March of 2009, I took my current post in Aberdeen.
Outside of work I spend a significant amount of time in the gym. Greco wrestling is my favourite and I have been lucky to find a place and group people in every country I have been to, to continue practicing it in some form and shape. The gym had to be replaced by jogging and cycling during COVID-19.
- Research
-
Research Overview
Skeletal muscle is responsible for many vital functions, including locomotion, respiration, maintenance of glucose homeostasis, thermoregulation, protection of bones and internal organs, and it also serves as a source of amino acids in time of starvation. The average appendicular muscle mass in healthy middle age females and males is 20 kg and 30 kg, respectively. However, individual differences are substantial; same sex, similar age and height people can differ 2-fold in muscle mass. The proportion of the fast-to-slow twitch fibres in the muscle can also differ from 20/80% to 80/20%. These differences impact on strength, power, and endurance of muscle contractions and are also associated with the risk of diseases which rely on muscle function. In adolescence and young adults, differences in muscle properties can influence success in sports, but it becomes increasingly important later in life. Low muscle mass and function increase the risk of type 2 diabetes and, compounded by aging-related decline, leads to frailty and admissions of the elderly to long-term care facilities. These differences in muscle properties result from the effects of developmental and postnatal environment, and from the genetic influences that can affect both pre- and post-natal phases. My research focus is to understand the physiological and genetic mechanisms underlying individual differences in skeletal muscle. This can lead to better prevention and discovery of new targets for pharmacological interventions to reverse the loss of muscle mass and function.
Research Areas
Research Specialisms
- Genetics
- Physiology
- Biomedical Sciences
Our research specialisms are based on the Higher Education Classification of Subjects (HECoS) which is HESA open data, published under the Creative Commons Attribution 4.0 International licence.
Current Research
My research aims to identify the genetic causes underlying the differences in muscle mass. We have carried out and integrated genome-wide association analyses in humans, using the UK Biobank resource, and in the laboratory mouse and rat models. The main outcome of these analyses were 182 genomic loci associated with differences in appendicular muscle mass in middle age, healthy individuals (Figure 1).

Figure 1. Map of Genome Associations with the Appendicular Muscle Mass in Humans. Significance level is presented on the vertical axis, while the chromosomal position of each genetic marker is shown on the horizontal axis. The red line across the plot represents the genome-wide threshold of significance (p < 5 x 10-8). This plot shows the association of variants with minor allele frequency >0.001 (Hernandez Cordero et al. 2019).
Furthermore, we found that inheriting large number of the “reducing” variants across these loci leads to lower muscle mass in the elderly population (Figure 2), indicating a sustained influence of those genetic variants across the lifespan. We also discovered that around a dozen of these loci overlap the syntenic regions of mouse and rat genomes. Syntenic regions refer to blocks of homologous genes arranged in the same order in the genome of different species. This convergence on the syntenic regions implicate a common cause and mechanism of function across mammalian species.
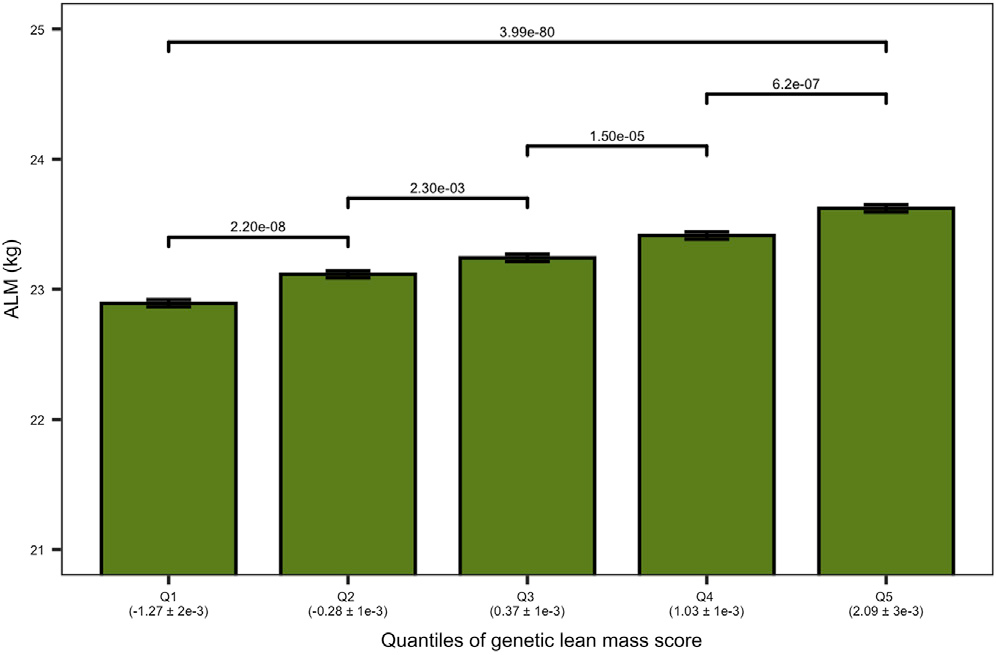
Figure 2. Genetic Lean Mass Score Affects the Appendicular Lean Mass (ALM) in Elderly Humans. The plot shows the ALM (kg) of the elderly cohort on the vertical axis. The elderly cohort was ranked by “genetic lean mass score” (calculated as a sum across 182 loci of the products of locus effect size and genotype dosage) and clustered in five quantiles (Q1–Q5) (horizontal axis). The average genetic lean mass score (±standard error) of each quantile is shown in parentheses below the horizontal axis. The overall quantile effect of the genetic lean mass score on ALM was tested with the Kruskal-Wallis test, and the resulting p-value is presented on the horizontal line above the bars. The ALM median differences between the groups were tested using a Wilcoxon test; the significance level of each comparison is presented above the horizontal lines with a Holm adjusted p-value (Hernandez Cordero et al. 2019).
The ultimate goal of genome-wide association studies is to identify the causative genes and understand mechanism of their effects. Achieving this is challenging because the confidence interval of each identified locus contains more than one, and often dozens, of genes. Furthermore, an interesting finding of these studies was that very few identified loci harbour genes known to affect myogenesis or growth and maintenance of muscle tissue. This means two things: 1) novel genes affecting skeletal muscle will be identified, and 2) additional studies are needed in order to prioritise and validate causative genes. We are currently working on validation and understanding the role of STC2 and POU3F4 encoding genes.
In a recent study we found that STC2 acts as a supressor of muscle growth (Lionikas et al. 2023). Mice lacking the Stc2 gene had up to 10% larger muscles than their wildtype littermates (Figure 3). This increase was mediated by larger cross-sectional area of muscle fibres. The STC2 induces this effect by supressing the insulin-like growth factor signalling which promotes protein accrual in skeletal muscle tissue.

Figure 3. Stc2 effects on muscle weight. The knockout mice (KO) have larger tibialis anterior (TA), externsor digitorum longus (EDL), plantaris (Plant) and soleus muscles. Sex-adjusted weight shown. Red symbols - female, black - male. The horizontal line represents the mean value, the error bars represent standard deviation (modified from Lionikas et al. 2023).
Past Research
Muscle mass can differ for two main morphological reasons: the number of muscle fibres comprising the muscle and the size of the fibres. The fibre number in mammals is determined prenatally and very shortly after birth and appears to be stable until some fibres are lost due to aging. The fibre size, i.e. cross-sectional area, is very pliable and depends on environmental factors such level and type of physical activity, constitutive factors such as sex and age, and genetic factors. Assessing the contribution between the fibre number and fibre size is important because they are influenced at different phases of the lifespan, and therefore likely to be determined by different genetic and molecular mechanisms. However, assessment of the role between the number and size of muscle fibres is not practical in humans due to an invasive nature of the biopsy procedure. We therefore conducted studies in laboratory mice. We studied transverse sections of mouse soleus muscle because a) all the fibres present in the muscle can be captured in one 10-µm thick slice; and b) unlike most rodent muscles, the soleus is similar to human muscles with respect to the expressed fibre types, i.e. type I and type IIA, and their proportions. Similarly to humans, muscle mass can differ more than 2-fold between inbred mouse strains (Figure 3). A difference between inbred strains housed under the same environmental conditions is an outcome of inheritance of the variants of genes influencing skeletal muscle. These studies revealed a number of strains differing in muscle mass. Importantly, in some of the strains, e.g. BEL and BEH, this difference is due to both the number and size of muscle fibres (Figure 3), but others, e.g. LG/J and SM/J, differ in the size but not the number of fibres (Lionikas et al. 2010, Carroll et al. 2011). These findings indicate that: 1) BEH and BEL strains captured variants of the genes involved both in the developmental processes and the postnatal growth and maintenance of the muscle, and 2) the LG/J and SM/J strains captured genetic variants involved only in muscle growth and maintenance. We later conducted a genome-wide association analysis on muscle weight of mice derived from a LG/J and SM/J intercross and found several genomic regions contributing to the difference in fibre size between these two strains (Gonzales et al. 2018).
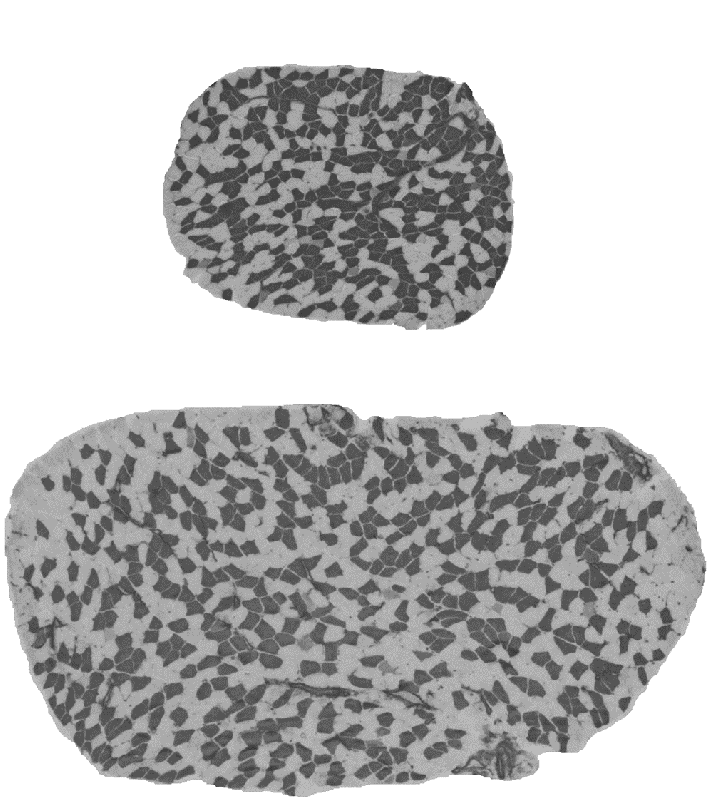
Figure 3. Cross-sections of soleus muscle from BEL (top image) and BEH (bottom image) strains. Samples were taken from male mice of the same age, subjected to ATPase staining following acid preincubation and imaged with 5X objective. Dark fibres are type I, pale fibres IIA. There are over 2-times more fibres and the fibres are over 2-times lager in the BEH strain soleus compared to BEL.
Knowledge Exchange
Skeletal muscle is a source of our strength, power, and stamina. Muscles enable us to move and breathe, protect our bones and internal organs, help to maintain healthy level of blood sugar, and generate heat to regulate body temperature. However, we become weaker as we age. What happens to our muscles? This animation, entitled "Ageing muscle: use it or lose it", explains the workings of the muscles, how muscles change as we age, and what can be done to maintain them throughout our lifespan.
Video powered by life-science-animation.com
Collaborations
Dr David Blizard, The Pennsylvania State University
Dr Abraham Palmer, University of California San Diego
Dr Veronique Blanquet, University of Limoges
Dr Thomas Coate, Georgetown University
Dr Claus Oxvig, Aarhus University
Funding and Grants
2007 – 2009 Dissection of Muscle Weight QTL via Congenic Strains (R03 AR052879); National Institute of Arthritis Musculoskeletal and Skin Diseases ($145,000 total cost); Role: PI
2009 - 2014 Genetic Variation of Muscle Mass (R01 AR056280); National Institute of Arthritis Musculoskeletal and Skin Diseases ($1,326,999 total cost); Role: Co-I
2009-2013 Genetic Mechanisms of Muscle Fibre Variation; European Commision – Marie Curie International Reintegration Grant (€87,500 total cost); Role: PI
- Publications
-
Page 1 of 6 Results 1 to 10 of 52
Differential Organ Ageing Is Associated With Age-Related Macular Degeneration
Aging Cell, vol. 24, no. 5, e14473Contributions to Journals: Articles- [ONLINE] DOI: https://doi.org/10.1111/acel.14473
- [OPEN ACCESS] http://aura.abdn.ac.uk/bitstreams/1e0a7d19-62ee-447b-93f3-e13470e4a708/download
- [ONLINE] View publication in Scopus
Generative AI workshops to enhance skills and confidence in academic practice
Annual Academic Development Symposium 2025Contributions to Conferences: PostersValidation of positional candidates Rps6ka6 and Pou3f4 for a locus associated with skeletal muscle mass variability
G3: Genes, Genomes, Genetics Mission, vol. 14, no. 5, jkae046Contributions to Journals: ArticlesStanniocalcin-2 inhibits skeletal muscle growth and is upregulated in functional overload-induced hypertrophy
Physiological reports, vol. 11, no. 15, e15793Contributions to Journals: ArticlesAnalysis of independent cohorts of outbred CFW mice reveals novel loci for behavioral and physiological traits and identifies factors determining reproducibility
G3: Genes, Genomes, Genetics Mission, vol. 12, no. 1, jkab394Contributions to Journals: ArticlesGenome-wide Associations Reveal Human-Mouse Genetic Convergence and Modifiers of Myogenesis, CPNE1 and STC2
American Journal of Human Genetics, vol. 105, no. 6, pp. 1222-1236Contributions to Journals: Articles- [ONLINE] DOI: https://doi.org/10.1016/j.ajhg.2019.10.014
- [OPEN ACCESS] http://aura.abdn.ac.uk/bitstreams/1128a8e4-06b2-4ded-9190-0995cbb18532/download
- [ONLINE] View publication in Scopus
Myostatin dysfunction is associated with lower physical activity and reduced improvements in glucose tolerance in response to caloric restriction in Berlin high mice
Experimental Gerontology, vol. 128, 110751Contributions to Journals: Articles- [ONLINE] DOI: https://doi.org/10.1016/j.exger.2019.110751
- [ONLINE] View publication in Scopus
Myostatin dysfunction does not protect from fasting-induced loss of muscle mass in mice
Journal of musculoskeletal & neuronal interactions, vol. 19, no. 3, pp. 342-353Contributions to Journals: ArticlesLow Citrate Synthase Activity Is Associated with Glucose Intolerance and Lipotoxicity
Journal of Nutrition and Metabolism, vol. 2019, 8594825Contributions to Journals: Articles- [ONLINE] DOI: https://doi.org/10.1155/2019/9153809
- [OPEN ACCESS] http://aura.abdn.ac.uk/bitstreams/f7d912a6-02be-4019-b826-e637741bbb27/download
- [ONLINE] View publication in Scopus
- [ONLINE] View publication in Mendeley
- [ONLINE] Corrigendum
Genome wide association analysis in a mouse advanced intercross line
Nature Communications, vol. 9, 5162Contributions to Journals: Articles
